Discovering Novel RNA Isoforms in Mouse Brown Fat
My bioinformatics pipeline/technical demo to uncover new RNA transcript variants using long-read RNA sequencing and as a learning tool.
Project Summary
This project analyzes a nanopore long-read RNA-seq dataset from murine (brown) brown adipose tissue
using a reproducible FLAIR-based pipeline. I identified both known and previously unannotated (novel) isoforms and quantify their expression.
This project also serves as a personal learning tool for an introduction to bio-informatics, focusing on basic data-analysis/pipeline.
Dataset
- Source: NCBI SRA
- ID and Link: SRR33470049
- Technology: Oxford Nanopore long-read sequencing (on GridION)
- Tissue: Mouse brown fat
- Reference/Annotations: Ensembl FTP
Pipeline Overview
- Environment Setup (Python 3.12, dependency installation, setup working env and directory)
- Quality Control (NanoPlot)
- Read Alignment (minimap2)
- Transcript Correction (FLAIR, not the NLP one)
- Transcript Collapsing
- Quantification of Isoforms
- Analysis & Visualization
Results
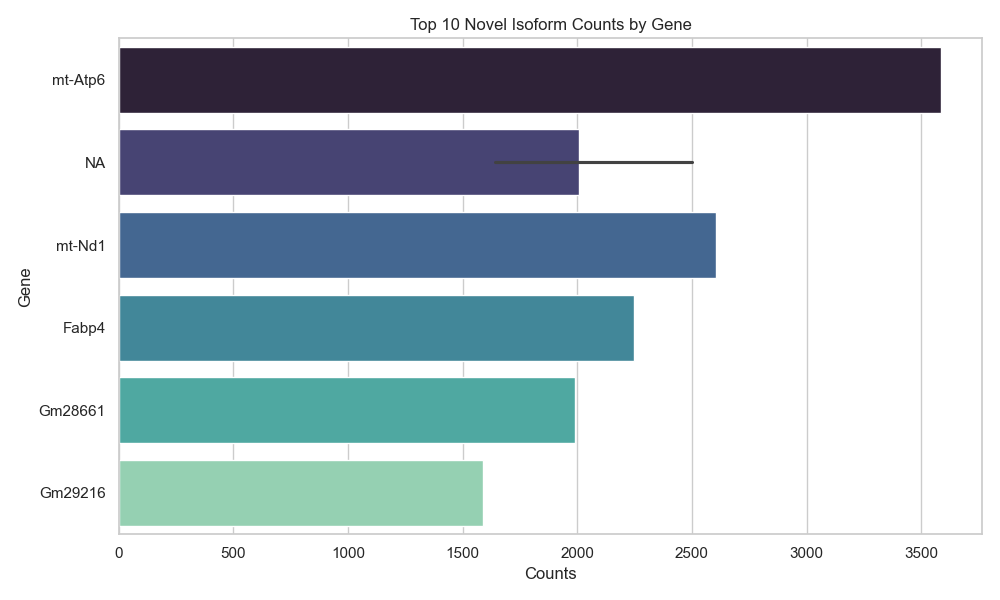
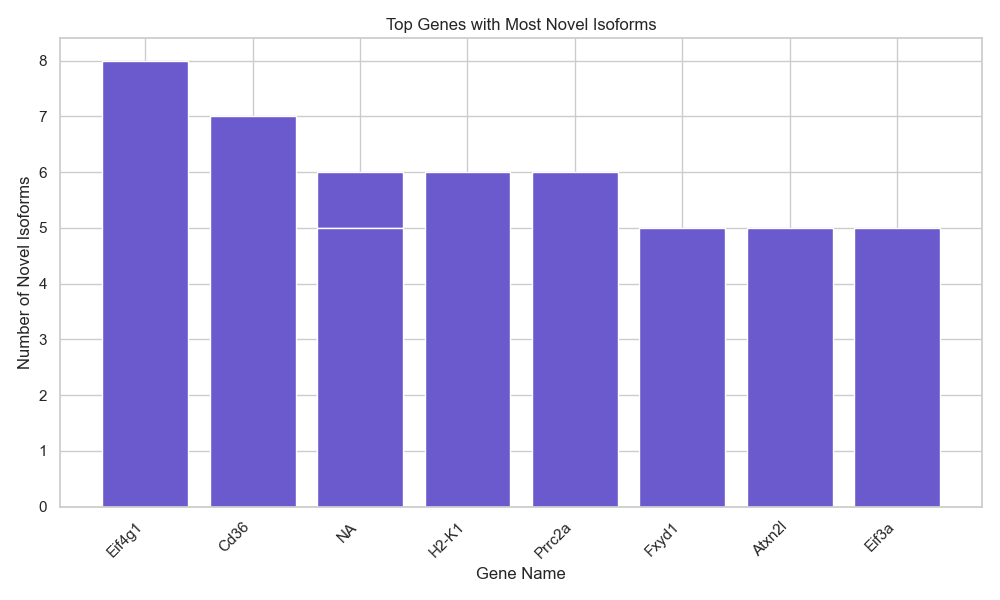
- 11,394 novel isoforms identified
- Highly variable expression patterns across genes
- Top novel isoform genes:
Eif4g1, Cd36, H2-K1
- Established a decent pipeline for data-analysis and learned basic genomics concepts while doing it! :)
These results highlight previously unknown RNA diversity (with some admitted limitations) in metabolically active brown fat tissue.
Limitations
- This project does focus more as a learning data-analysis pipeline for python, so I'll admit there are limitations and this is quite limited.
- I only used one sample and did not compare to publicly available RefSeq/GENCODE or any white papers, this was more of a tech demo.
- My reference could just be rough/incomplete.
- Only looked at specifically brown mouse fat.
- Low sensitivity could have been a possibility.
- I didn't really look into the context of the study, so not sure of the exact environmental context for these lil mice.
Why It Matters
Identifying novel isoforms can uncover new mechanisms of gene regulation, alternative splicing, and tissue-specific gene expression — especially in dynamic tissues like brown fat, which plays a role in energy metabolism.
Repository
Explore the code, data prep, and full Jupyter notebook with more information/indepth:
View on GitHub
About me!
I'm Thaddeus Lipke, a SWE/EMT-B certified grad from Columbia Uni who wanted to learn some bio-informatics and get more into python.
Feel free to contact me, I'd like to learn more if something needs clarification or corrections (please correct me)! My contact is on the github repo for this!
This project helped me learn genomics tools like FLAIR, minimap2, and samtools — and apply them to a real-world RNA sequencing problem with a mouse dataset.

